BME reports on our COVID-19-focused research
BME University of Technology and Economics reports on our COVID-19-focused research (in Hungarian)
http://www.bme.hu/hirek/20200421/A_jarvanyhoz_kapcsolodo_gyogyszerkutatast_segitik_a_BME_feherjeszerkezeti_vizsgalatai
„A JÁRVÁNYHOZ KAPCSOLÓDÓ GYÓGYSZERKUTATÁST SEGÍTIK A BME FEHÉRJESZERKEZETI VIZSGÁLATAI”
2020. április 21.
A legújabb műegyetemi kutatásról, a COVID-19 tulajdonságairól, a lehetséges gyógymódokról, és a teljes immunitás megszerzésének esélyéről is beszélgettünk a VBK kutatónőjével.
„Tudósokként minél szélesebb körű ismereteket kell szereznünk a láthatatlan ellenségnek számító új koronavírusról, magánemberként pedig az a feladatunk, hogy a személyes kontaktusok számának minimalizálásával időt nyerjünk az akadémiai szakembereknek a gyógymód megtalálásához” – fogalmazta meg a tudományos kutatók és a lakosság felelősségét a járványügyi vészhelyzettel összefüggésben a bme.hu-nak Vértessy Beáta, a BME Vegyészmérnöki és Biomérnöki Kar (VBK) Alkalmazott Biotechnológia és Élelmiszertudományi Tanszék tanszékvezető egyetemi tanára, aki a Műegyetemen szerkezeti biológiai, míg a Természettudományi Kutatóközpont (TTK) Enzimológiai Intézetében enzimológiai (azaz az enzimek működésével kapcsolatos) kutatásokat folytat. Hozzátette: az idő előrehaladtával és a fertőzöttek kórtörténetének megismerése által nyert értékes információk rendszerezésével a betegségről egyre többet tudhatunk meg.

A BME kutatónője beszámolt arról is, hogy a Műegyetem részt vesz a vírus megismerésének tudományos folyamatában. A VBK-n az új koronavírus (SARS-CoV-2) biotechnológiai módszerekkel előállított RNS polimeráz enzimének modellezésén, valamint e fehérje háromdimenziós térszerkezetének megalkotásán dolgoznak. Az RNS polimeráz enzim felelős a vírus szaporodásáért, úgy hogy katalizálja a virális RNS sokszorosítását. A szerkezet megismerésével válik lehetővé gyógyszerjelölt molekulák azonosítása. „Ez a lépés a gyógyszerfejlesztés egyik előfeltétele. Normál esetben akár évekig is eltarthat új termékek fejlesztése és engedélyezése, ám a világjárvány elleni küzdelem kiemelkedő jelentőséggel ruházta fel a virológiai vizsgálatokat, így jelenleg olyan gyógymódokon is dolgoznak, ahol már korábban engedélyezett gyógyszereket vetnek be a koronavírus okozta betegség ellen. Ez új lendületet és lehetőséget ad a műegyetemi szakembereknek is” – emelte ki Vértessy Beáta. A BME-n évek óta a hazai és a nemzetközi fehérjeszerkezet-kutatások élmezőnyébe tartozó vizsgálatok zajlanak több területen is, az intézményben dolgozó műegyetemi szakemberek az eddigi tapasztalatukat és tudásukat felhasználva a pandémia idején minden tőlük telhetőt meg fognak tenni a járvány leküzdéséért.
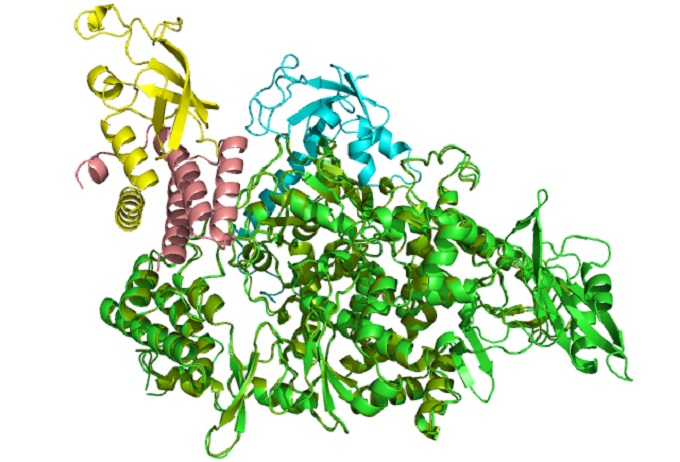
Az ábra a jelenlegi járványt okozó SARS-CoV-2 koronavírus szaporodásáért felelős RNS polimeráz enzim modelljét mutatja (zöld színű szalagmodell), több virális segítő fehérjével együtt (sárga, kék rózsaszínű szalagmodellek).
A műegyetemi oktató-kutató ismertette, hogy a koronavírus örökítőanyaga nem a „tipikus” DNS (dezoxiribonukleinsav), hanem az RNS (ribonukleinsav): a vírus a fertőzött sejtben a saját RNS-ét szaporítja, a virális RNS polimeráz által. További fontos jellemzője ennek a koronavírusnak az, hogy RNS molekulája egyben azonnal hírvivő RNS-ként is szolgál, amiről a virális fehérjék képződnek. Az RNS a DNS-el szemben könnyebben mutálódik, kémiailag aktívabb, így egyben kevésbé védett a környezeti hatásokkal szemben. A most járványt okozó koronavírus evolúciós rokonságot mutat több egyéb betegséget okozó vírussal. Vannak például olyan koronavírusok, amelyek a közönséges nátha egyes típusaiért felelősek, továbbá hasonló alapon működnek a sárgaláz, a ZIKA láz és a hepatitis C betegségek vírusai. „Az új koronavírus elleni fellépéskor a szakemberek megvizsgálják a rokon jellegzetességeket mutató kórokozókkal szemben sikeresnek bizonyult gyógyszerek potenciális bevethetőségét is. Ezen termékek többsége nagy valószínűséggel nem alkalmazható teljes sikerrel a mostani járványban, azonban enyhe vagy közepesen erős tünetek esetén képesek lehetnek az enyhítésre, emellett a betegség időbeli lefolyását is rövidíthetik” – mutatott rá a járvány időszakában alkalmazható antivirális készítmények fontosságára Vértessy Beáta.
„A rendkívül súlyos betegségeket és halált is okozó új koronavírus (SARS-CoV-2 vírus) veszélyességének egyedisége részben abban nyilvánul meg, hogy nagyobb hatékonysággal fertőz, mint a korábbi SARS koronavírus. Gyakori, hogy a fertőzés nem jár tünetekkel, de a fertőzött személy képes továbbadni a vírust másoknak, így a betegség gyorsan terjedhet. A vírus kópiái egy fertőzött emberben rövid időn belül sok új sejtet fertőznek meg. A kórokozó vírus felszínén található jellegzetes tüske formájú fehérjék az ACE2 nevű emberi sejtfelszíni membránfehérjéhez kötődnek, és ezután tud a vírus RNS-e bejutni az emberi sejtbe. Az ACE2 légutak és a tüdőhólyagocskák bőrfelszíni sejtjein megtalálható, így ezek a sejtek gyorsan károsodnak. A járvány elleni küzdelem egyik lehetősége azt célozza, hogy speciális módszerrel megakadályozzuk ezt a fajta kapcsolódást” – részletezte az egyik gyógymód-alternatívát Vértessy Beáta. Ennek során mintegy csaliként készülnek a kutatók olyan anyagot bevetni, amelyhez a kórokozó vírus tüskéi hozzákötődnek, így nem tudnak kapcsolatot létesíteni az emberi sejt felszínén lévő ACE2 fehérjével.
A BME kutatónője kifejtette, hogy az új koronavírus eltérő intenzitással és tünetekkel fertőz, emiatt a kórokozóval szembeni hatékony küzdelem a személyre szabott diagnosztikában és terápiában rejlik. Az eltérő emberi reakciók hátterében az életkor és a meglevő egészségi állapot mellett bizonyos mértékben genetikai sajátosságok is állhatnak, vélhetően ez részben magyarázza azt a jelenséget, hogy egyesek miért reagálnak sokkal érzékenyebben a betegség kialakulásakor, míg másoknál tünetek sincsenek. A genetikai okok feltárására már egy, a Helsinki Egyetem Molekuláris Orvostudományi Intézete által összefogott kutatást indítottak a szakemberek. Vértessy Beáta szerint a genetikai sokszínűség feltérképezése segítheti a betegség kialakulásának jobb megértését. A lehetséges terápiák sorában egyébként felmerült egy további alternatíva is, amelynek során a gyógyult betegektől nyert vírusellenes ellenanyagokat tartalmazó vérplazmát alkalmazzák. Ez egyelőre csak ideiglenes megoldásnak bizonyul, de az ellenanyagok felhasználásának további lehetőségeit (pl. biotechnológiai alapú előállítás) Magyarországon is vizsgálják.
Az USA-ban önkénteseken tesztelnek vírusellenesnek vélt vakcinákat, ám e szerek engedélyeztetése az emberi egészség védelmében meghozott előírások szerint egy hosszadalmas folyamat, még hónapokig eltarthat”. Magyar kutatók érdeme egy friss tudományos eredmény, amely a COVID-19 betegség elleni küzdelemben is jelentős előrelépés lehet: az MTA-SE Lendület Diabétesz Kutatócsoport több éve foglalkozik a számos sejtben megtalálható szigma-1 receptor megismerésével, amely gátolja a szervezeten belüli hegesedési folyamatokat (fibrózist). Ilyen hegesedés következik be a koronavírussal összefüggésbe hozható – a páciens számára sok esetben végzetes kimenetellel járó – tüdőgyulladás folyamán. A kutatók most azt elemzik szabadalmukban, hogy a tüdőgyulladás ellen hogyan lehet hatékonyan fellépni a szigma-1 receptor segítségével. Új felvetés az is, hogy fluvoxamin tartalmú, már engedélyezett és piacon lévő antidepresszánsok gátolhatják a tüdőgyulladás okozta hegesedést.
Vértessy Beáta a fertőzöttek életkori sajátosságairól is beszélt: „a halálesetek jelentős részét az idősek körében az alapbetegségek erősítő hatása mellett a súlyos tüdőgyulladás okozza, amelyért az immunrendszer fertőzéssel szembeni 'túlreagálása' a felelős, ez az ún. 'citokin vihar'. Hozzáfűzte: „ugyanez a helyzet fordulhat elő például az autoimmun betegségekkel szenvedőknél, ám a gyermekeknél még nincs meg ez a túlzott visszacsatolás. Jelenlegi ismereteink szerint a nagyon fiatalok is elkapják a vírust, de bennük csak nagyon ritkán alakul ki súlyos betegség a koronavírus miatt. Összességében elmondható, hogy az egészséges emberek esetében a kor előrehaladtával nő a tünetek súlyossága, és egy új vizsgálat szerint nemcsak a tüdőt, hanem a vesét, a szívet és a keringést is roncsolja a kórokozó” – hangsúlyozta Vértessy Beáta.
A professzor asszony szerint egyelőre nincs arra garancia, hogy az egyszeri megfertőződés örök védettséget ad az új koronavírussal szemben. „A jól működő szervezet biztosan mutat valamiféle ellenállást, de mint minden más kórokozó, ez is idővel mutálódhat, mint ahogyan ez már megtörtént, így a teljes immunitás megkérdőjelezhető. Jelenleg 8, jól elkülöníthető törzsét ismerjük az új koronavírusnak, amelyek szerencsére nagyon hasonló tulajdonságokat mutatnak, így a védekezésben könnyebb megtalálni a közös nevezőt az ellenszer (akár gyógyszer, akár vakcina) kifejlesztésekor.” Magyarországon több variáns jelenlétét is észlelték. „További súlyos bonyodalmat jelent azonban, hogy egy hordozó szervezetében akár többféle törzs vírusai is találkozhatnak, rekombinálódva mutálódhatnak, így az illető már egy jelentősen megváltozott kórokozót adhat tovább. Részben ezért olyan fontos a személyes kontaktusok minimalizálása és a fertőzés továbbadásának ily módon történő megakadályozása” – mutatott rá a kockázatokra a műegyetemi kutatónő.
Vértessy Beátát megkérdeztük a BCG-oltással kapcsolatos teóriákról is: „az Egészségügyi Világszervezet szerint egyelőre nincs bizonyítva, hogy a tuberkulózis megelőzésére használt vakcina a koronavírussal szemben bármilyen védettséget jelent. Ugyanakkor statisztikailag egyértelműen alátámasztott: azokban az országokban, pl. Magyarországon, ahol ez az oltás évtizedek óta kötelező, kevesebben betegedtek meg, és ami még fontosabb, alacsonyabb a halálozási arány”.
A bme.hu-nak adott interjúban arra kértük a Műegyetem oktató-kutatóját, értékelje a hazai járványügyi állapotot. Vértessy Beáta egyetértett azzal, hogy a személyes kontaktusok csökkentésére irányuló járványügyi óvintézkedések rendkívül fontosak, ugyanakkor hozzátette: „hazánkban átlagosan 300 ember hal meg naponta különböző okok miatt. Minden ilyen eset tragikus esemény, ám a dél-európai vagy az amerikai adatokhoz képest a Magyarországon koronavírussal összefüggésben elhunytak száma még mindig azt a képet mutatja, hogy hatékony a vírussal szembeni védekezés, mert eddig – az első új koronavírusos beteg március eleji diagnosztizálása óta – a koronavírus magyarországi áldozatainak aránya töredéke a szokásos napi halálozásnak. Sokkal inkább a fertőzöttséggel szembeni félelem és a bizonytalanság terhe nyomja az emberek lelkét, és tudomásul kell venni, hogy sajnos a végleges megoldásra egyelőre még várni kell”.
„A világjárvány után valószínűleg az új típusú koronavírus velünk marad: a parazita érdeke együtt létezni a gazdatesttel, ügyelve arra, hogy azt véglegesen ne betegítse meg, mivel egy, a mindkettőjük számára hosszú távon életképes 'szimbiózis' kialakítására törekszik. A kórokozó és az emberi test is e tanulófázis időszakában van most. Fontos megtanulnunk ellene a védekezést, illetve ha a vírus megszelídül, akkor a vele együttélést, mint ahogyan ez a történelem során számos egyéb hasonló esetben is lezajlott. Ebből a szempontból érdekes kiemelni, hogy sejtjeink örökítőanyagában, az emberi DNS genomban sok vírusból származó genetikai elem is található, amelyek korábbi fertőzések lenyomatai” – összegezte a professzor asszony.
TZS-GI
Fotó: Takács Ildikó















